ProteinDiscovery- Homology Modeling Softwares - |
|
|
Prediction of protein tertiary structures is an important process
in CADD (Computer Aided Drug Design). However, finding accurate
structural model is the most difficult task, and thus demands very
high skill and deep experience.
In-Silico Sciences Inc. introduces ProteinDiscovery software family, which were originally developed by Professor Hideaki Umeyama (School of Pharmaceutical Sciences, Kitasato University). The core softwares of the family are PDFAMS, homology modelling tools, with many optional modules. This software family offers you various functions and high resolution structural models that are comparable to the prediction by the most experienced experts. One of the successful outcome of the ProteinDiscovery is the protein structures of SARS (Severe Acute Respiratory Syndrome). In-Silico Sciences has started the world's first service to provide the protein structural data in order to promote the studies to develop drugs to treat the deadly respiratory illness and vaccines to prevent the infection. |
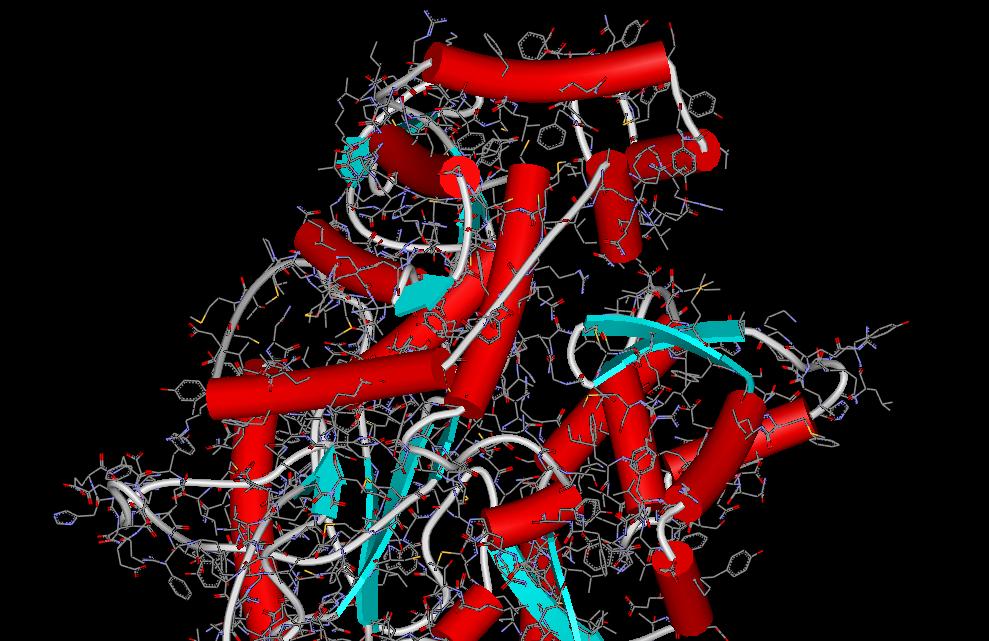 Tertiary Structure of the RNA Plymerase of SARS virus, obtained by PDFAMS softwares
|